Outline an approximate pairwise confidence region
ellipse.nls.RdThis function produces the ellipsoidal outline of an approximate pairwise confidence region for a nonlinear model fit.
Arguments
- x
The first argument should be an
nlsobject, usually resulting from a call tonls().- which
Which selects the pair of parameters to be plotted. The default is the first two.
- level
The confidence level of the region. Default 95%.
- t
The t statistic on the boundary of the ellipse.
- ...
Other
ellipse.defaultparameters may also be used.
Details
The summary function is used to obtain the approximate covariance matrix of the fitted parameters.
Examples
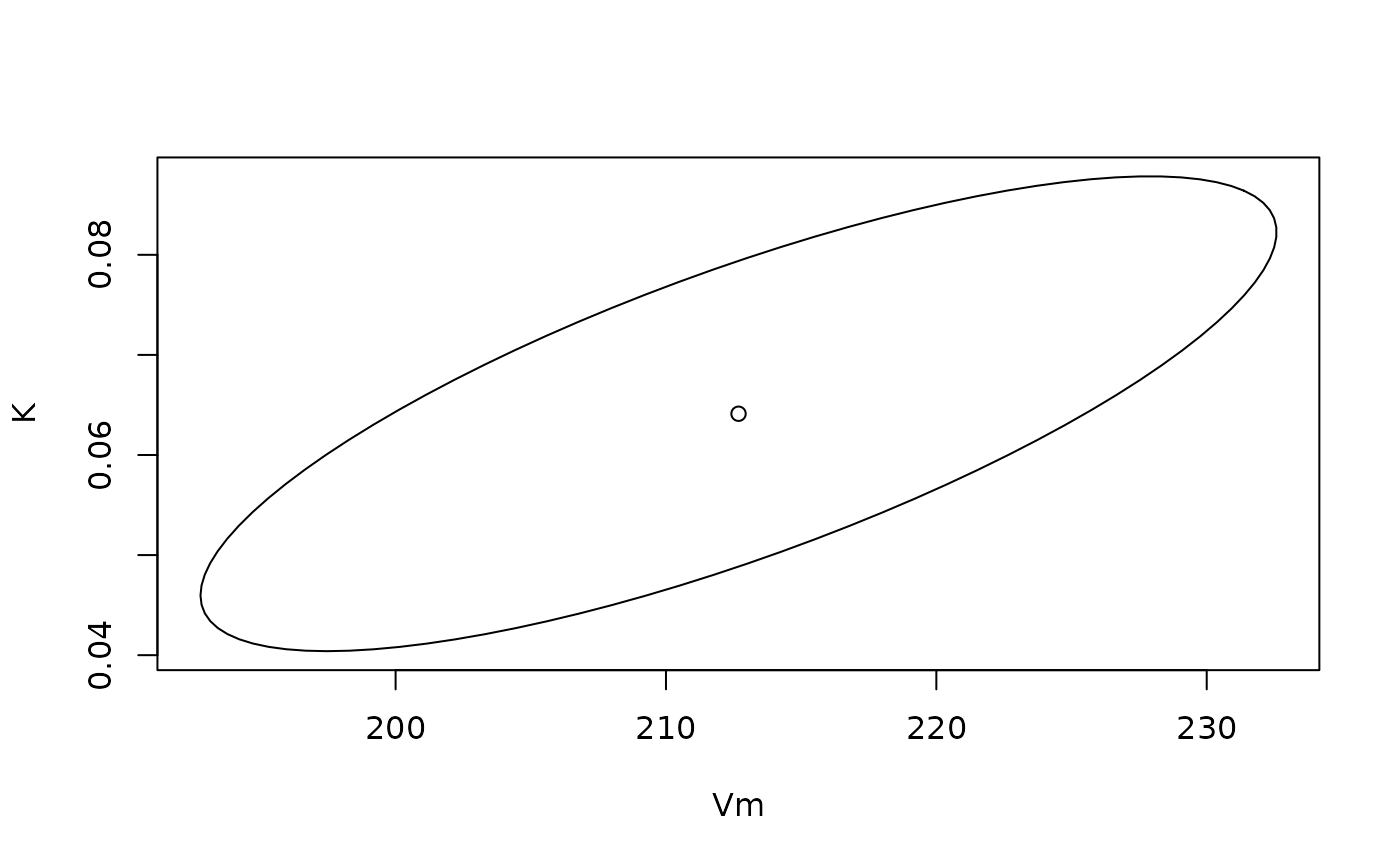
# Plot an approximate 95% confidence region for the weight and displacement
# parameters in the Michaelis Menten model
data(Puromycin)
fit <- nls(rate ~ Vm*conc/(K + conc), data = Puromycin, subset = state=="treated",
start = list(K = 0.05, Vm = 200))
plot(ellipse(fit,which=c('Vm','K')), type = 'l')
params <- fit$m$getPars()
points(params['Vm'],params['K'])