Pairwise profile sketch for GLM profiles
ellipse.profile.glm.RdThis routine approximates a pairwise confidence region for a glm
model.
Arguments
- x
An object of class
profile.glm.- which
Which pair of parameters to use.
- level
The
levelargument specifies the confidence level for an asymptotic confidence region.- t
The square root of the value to be contoured. By default, this is
qchisq(level, 2)for models with fixed dispersion (i.e. binomial and Poisson), and2 * qf(level, 2, df)for other models, wheredfis the residual degrees of freedom.- npoints
How many points to use in the ellipse.
- dispersion
If specified, fixed dispersion is assumed, otherwise the dispersion is taken from the model.
- ...
Extra parameters which are not used (for compatibility with the generic).
Value
An npoints x 2 matrix with columns having the chosen parameter names,
which approximates a contour of the function that was profiled.
Details
This function uses the 4 point approximation to the contour as described in Appendix 6 of Bates and Watts (1988). It produces the exact contour for quadratic surfaces, and good approximations for mild deviations from quadratic. If the surface is multimodal, the algorithm is likely to produce nonsense.
References
Bates and Watts (1988). Nonlinear Regression Analysis and Its Applications. Wiley. doi:10.1002/9780470316757 .
Examples
## MASS has a pairs.profile function that conflicts with ours, so
## do a little trickery here
noMASS <- is.na(match('package:MASS', search()))
if (noMASS) require(MASS)
#> Loading required package: MASS
## Dobson (1990) Page 93: Randomized Controlled Trial :
counts <- c(18,17,15,20,10,20,25,13,12)
outcome <- gl(3,1,9)
treatment <- gl(3,3)
glm.D93 <- glm(counts ~ outcome + treatment, family=poisson())
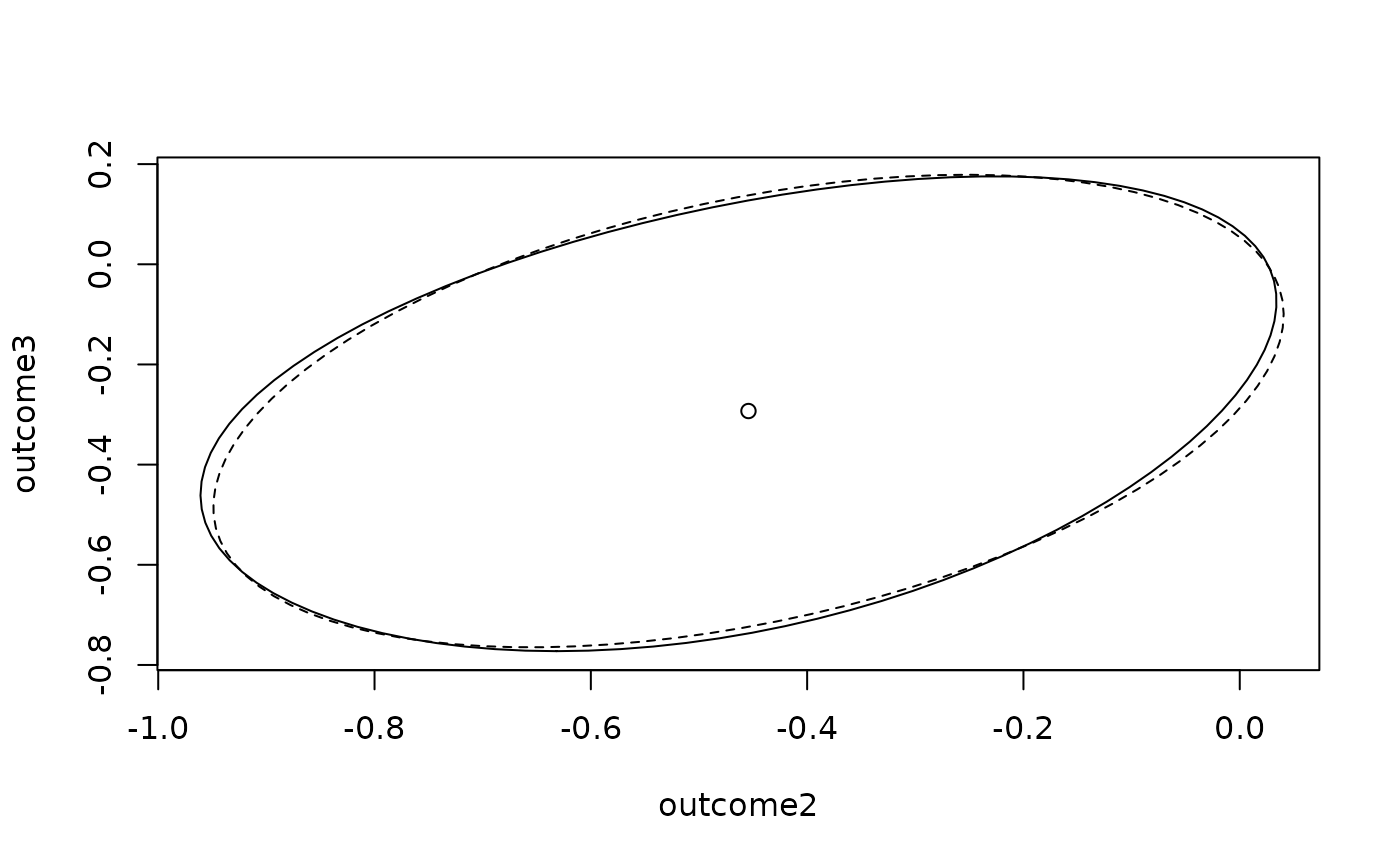
## Plot an approximate 95% confidence region for the two outcome variables
prof.D93 <- profile(glm.D93)
plot(ellipse(prof.D93, which = 2:3), type = 'l')
lines(ellipse(glm.D93, which = 2:3), lty = 2)
params <- glm.D93$coefficients
points(params[2],params[3])
 ## Clean up our trickery
if (noMASS) detach('package:MASS')
## Clean up our trickery
if (noMASS) detach('package:MASS')