Profile pairs
pairs.profile.RdThis function produces pairwise plots of profile traces, profile sketches, and ellipse approximations to confidence intervals.
Arguments
- x
An object of class
profile, generally the result of theprofile()function.- labels
The labels to use for each variable. These default to the variable names.
- panel
The function to use to draw the sketch in each panel.
- invert
Whether to swap the axes so things look better.
- plot.tau
Whether to do the profile tau (profile t) plots.
- plot.trace
Whether to do the profile trace plots.
- plot.sketch
Whether to do the profile sketch plots.
- plot.ellipse
Whether to do the ellipse approximations.
- level
The nominal confidence level for the profile sketches and ellipses.
- ...
Other plotting parameters.
Side Effects
Produces a plot on the current device for each pair of variables in the profile object.
Details
This function implements the plots used in Bates and Watts (1988) for nonlinear regression diagnostics.
Prior to ellipse version 0.5,
the pairs_profile function was a profile
method for the pairs generic. This caused
various conflicts, because graphics also exports a pairs
generic, and package MASS exported a profile
method for graphics::pairs. As of R version 4.4.0,
the MASS method will be in stats instead.
If x is a profile object then pairs_profile(x)
will call the function from this package. If you'd rather use
the MASS/stats method, then make sure the appropriate
package is loaded, and call pairs(x). (Prior to
ellipse 0.5, there were complicated rules to determine what
pairs(x) would do; those should still work for now, but
ellipse::pairs will disappear in a future release.)
References
Bates and Watts (1988). Nonlinear Regression Analysis and Its Applications. Wiley. doi:10.1002/9780470316757 .
See also
pairs, profile, ellipse.profile, ellipse.nls
Examples
# Plot everything for the Puromycin data
data(Puromycin)
Purboth <- nls(formula = rate ~ ((Vm + delV * (state == "treated"))
* conc)/(K + conc), data = Puromycin,
start = list(Vm = 160, delV = 40, K = 0.05))
Pur.prof <- profile(Purboth)
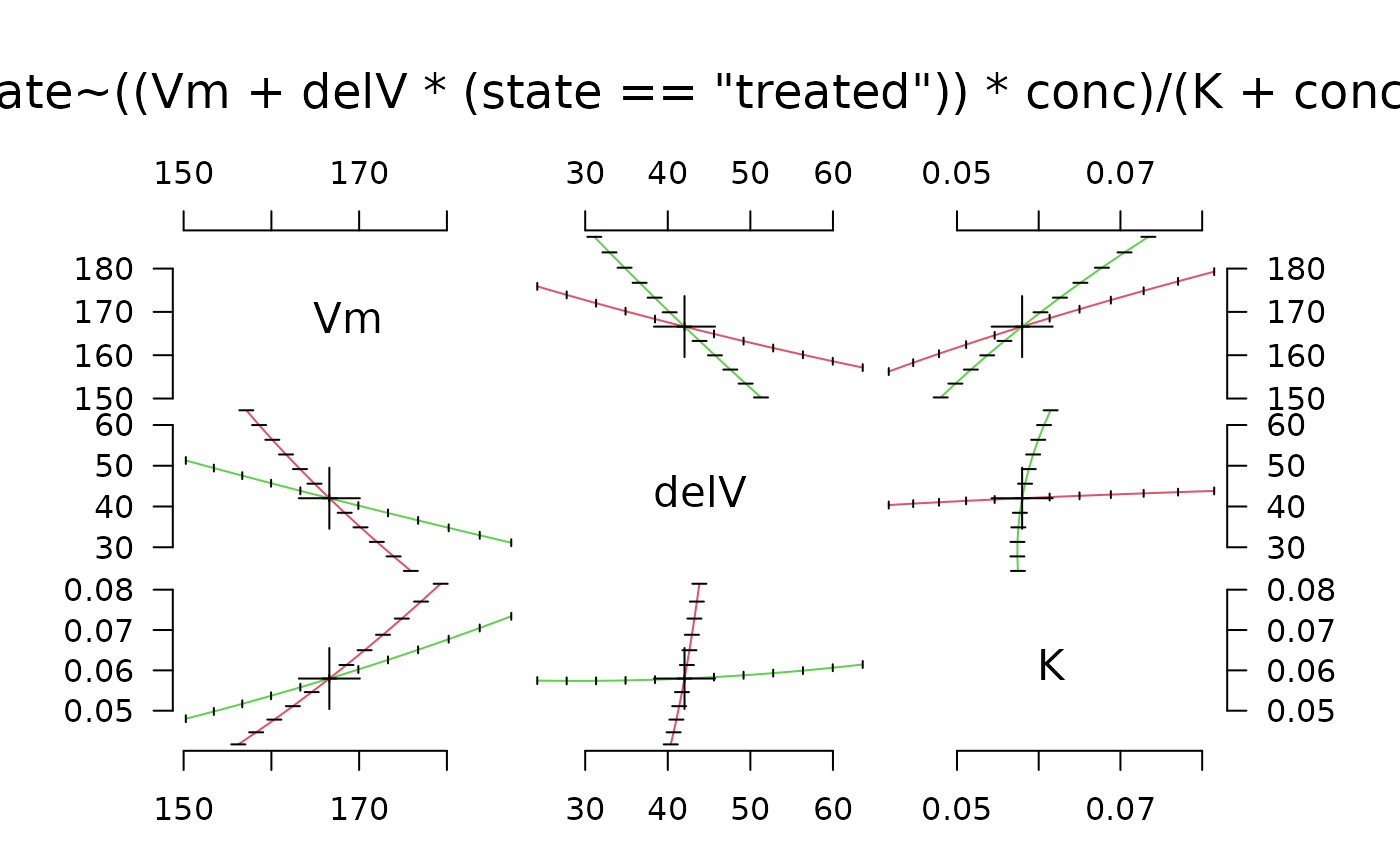
pairs_profile(Pur.prof, plot.ellipse = TRUE)
 # Show the corresponding plot from MASS/stats:
if (getRversion() < "4.4.0") {
loadNamespace("MASS")
} else
loadNamespace("stats")
#> <environment: namespace:MASS>
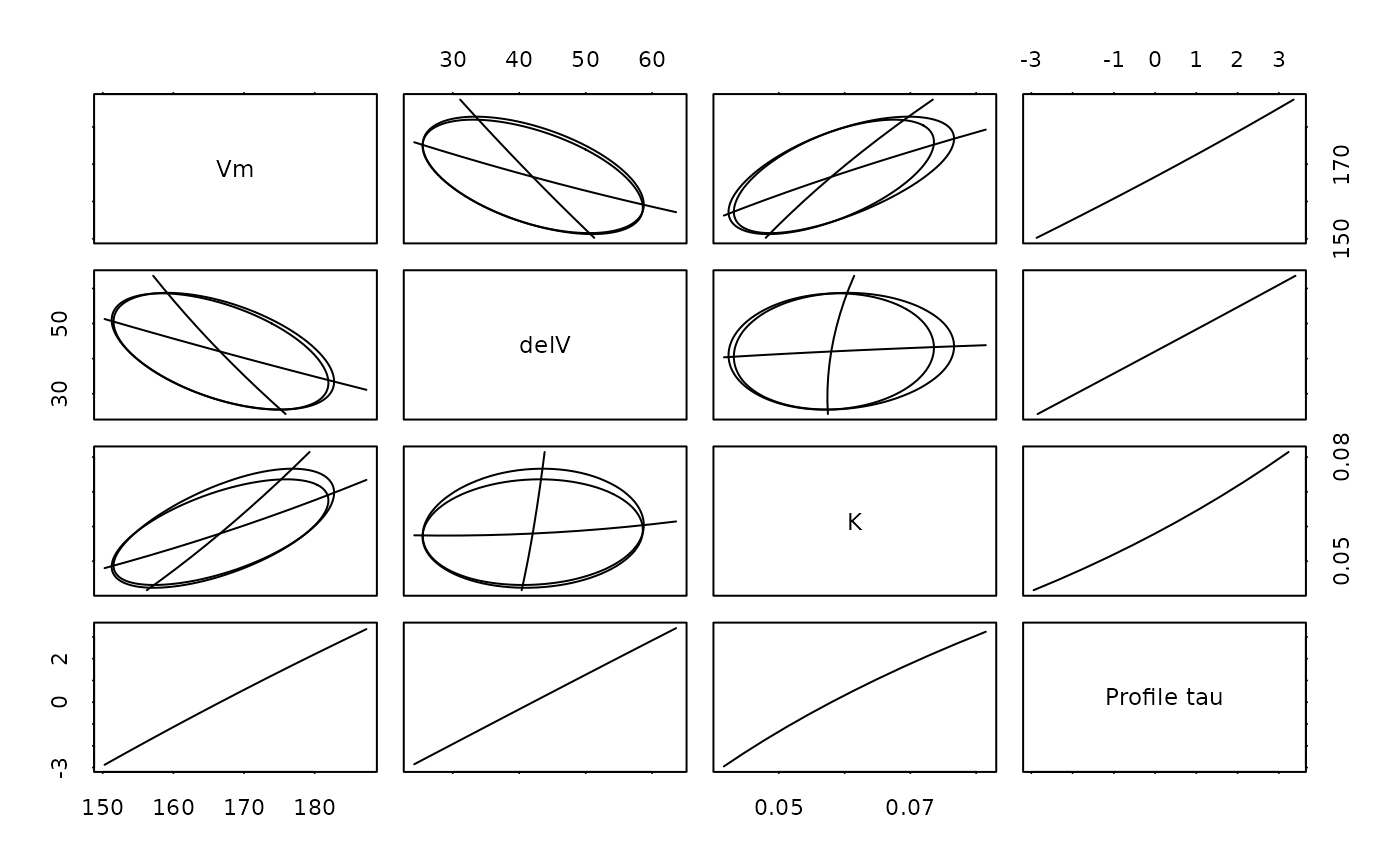
graphics::pairs(Pur.prof)
# Show the corresponding plot from MASS/stats:
if (getRversion() < "4.4.0") {
loadNamespace("MASS")
} else
loadNamespace("stats")
#> <environment: namespace:MASS>
graphics::pairs(Pur.prof)